pnetr: An R package for the PnET family of forest ecosystem models
Jun 9, 2025· ,,,,,·
1 min read
,,,,,·
1 min read
Xiaojie Gao (高孝杰)
Zaixing Zhou
Scott v. Ollinger
Jaclyn Hatala Mattes
Wenzhe Jiao
Jonathan R. Thompson
Abstract
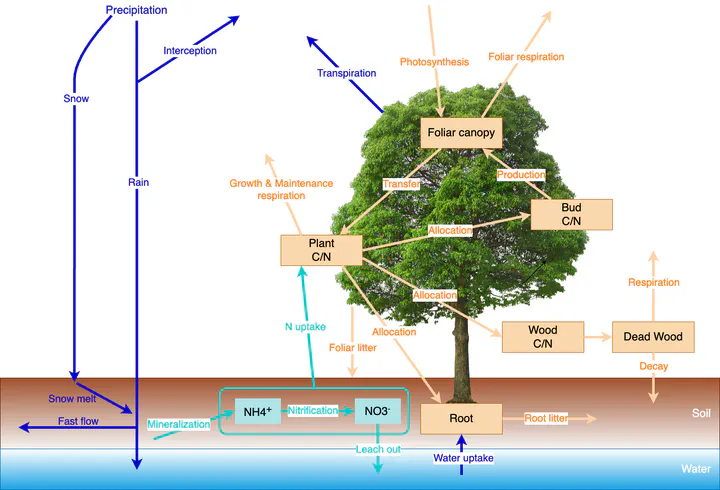
- Ecosystem models offer a rigorous way to formalize scientific theories and are critical to evaluating complex interactions among ecological and biogeochemical processes. In addition to simulation and prediction, ecosystem models are a valuable tool for testing hypotheses about mechanisms and empirical findings because they reveal critical internal processes that are difficult to observe directly. 2. However, many ecosystem models are difficult to manage and apply by scientists because of complex model structures, lack of consistent documentation, and low-level programming implementation. 3. Here, we present the ‘pnetr’ R package, which is designed to provide an easy-to- manage ecosystem modelling framework and detailed documentation in both model structure and programming. The framework implements a family of widely used PnET (net photosynthesis, evapotranspiration) ecosystem models, which are relatively parsimonious but capture essential biogeochemical cycles of water, carbon and nitrogen. We chose the R programming language because it is familiar to many ecologists and has abundant statistical modelling resources. We show- case examples of model simulations and test the effects of phenology on carbon assimilation and wood production using data measured by the Environmental Measurement Station (EMS) eddy- covariance flux tower at Harvard Forest, MA. 4. We hope ‘pnetr’ can facilitate further development of ecological theory and in- crease the accessibility of ecosystem modelling and ecological forecasting.
Type
Publication
Methods in Ecology & Evolution
- The open access article in Methods in Ecology & Evolution can be found here.
- There’s a blogpost describing the story behind this work also published on the journal website.
- The source code of the
pnetrpackage is available via GitHub. Although it’s an R package, we encourage users to modify the source code for testing the hypothetical processes of interest.